Exploratory Data Analysis
Code
library (gapminder)<- gapminder |> filter (year == 1982 ) |> rename (lifeexp = lifeExp, gdppercap = gdpPercap) |> select (country, lifeexp, continent, gdppercap) |> head () |> kable ()
Afghanistan
39.854
Asia
978.0114
Albania
70.420
Europe
3630.8807
Algeria
61.368
Africa
5745.1602
Angola
39.942
Africa
2756.9537
Argentina
69.942
Americas
8997.8974
Australia
74.740
Oceania
19477.0093
Code
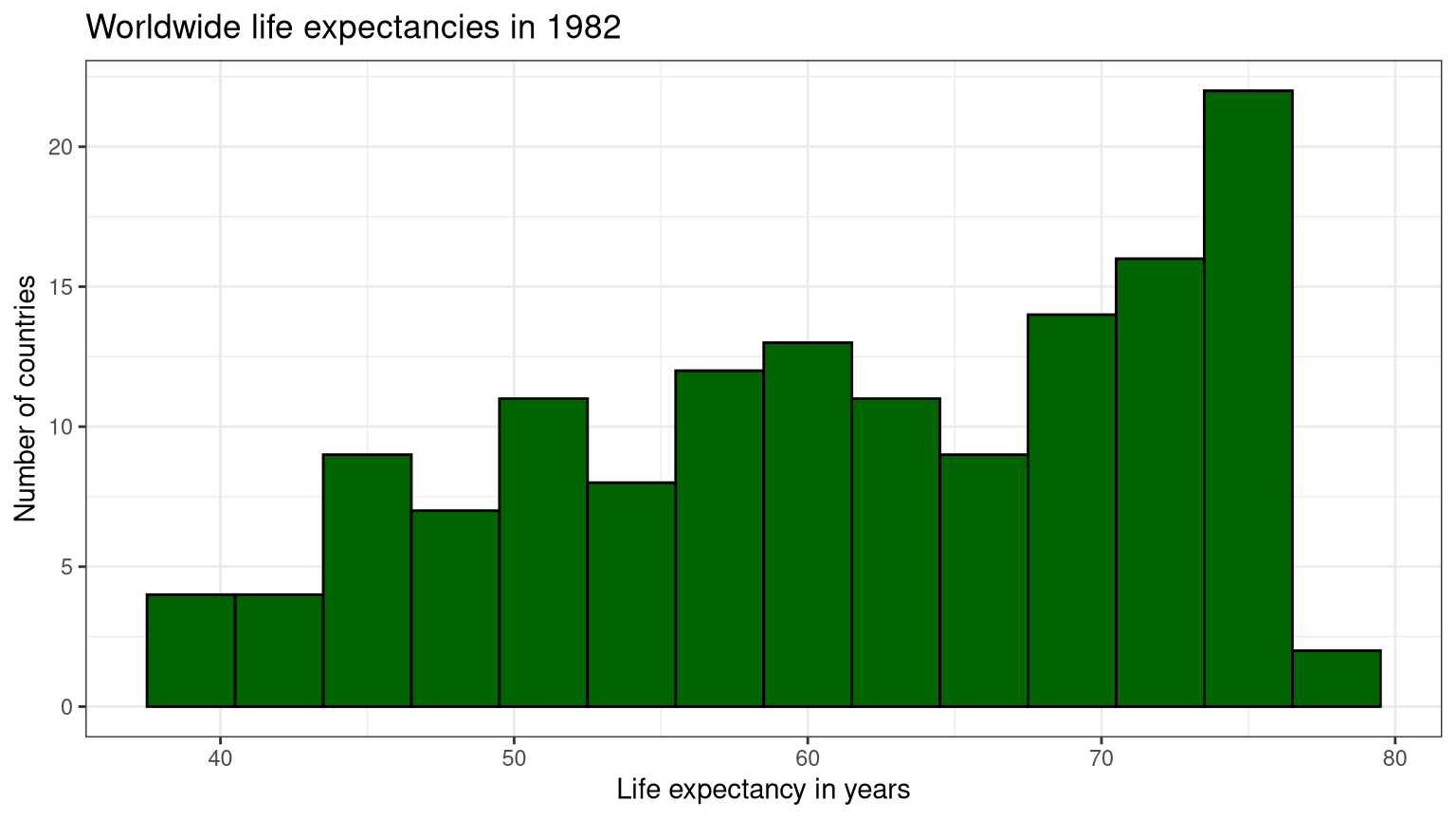
```{r} #| label: "fig-exp" #| fig-cap: "Worldwide life expectancies in 1982" ggplot (data = gapminder1982, aes (x = lifeexp)) + geom_histogram (binwidth = 3 ,color = "black" ,fill = "darkgreen" ) + theme_bw () + labs (title = "Worldwide life expectancies in 1982" ,x = "Life expectancy in years" ,y = "Number of countries" )```
Based on Figure 1 , woldwide life expectancies in 1982 has a unimodal skew left distribution with statistics given in Table 1 .
Code
```{r} #| label: "tbl-stat" #| tbl-cap: "Descriptive statistics" library (e1071)|> summarize (Median = median (lifeexp),IQR = IQR (lifeexp),Skew = skewness (lifeexp)-> T1|> :: kable ()```
Table 1: Descriptive statistics
62.4415
17.98125
-0.3427068
Code
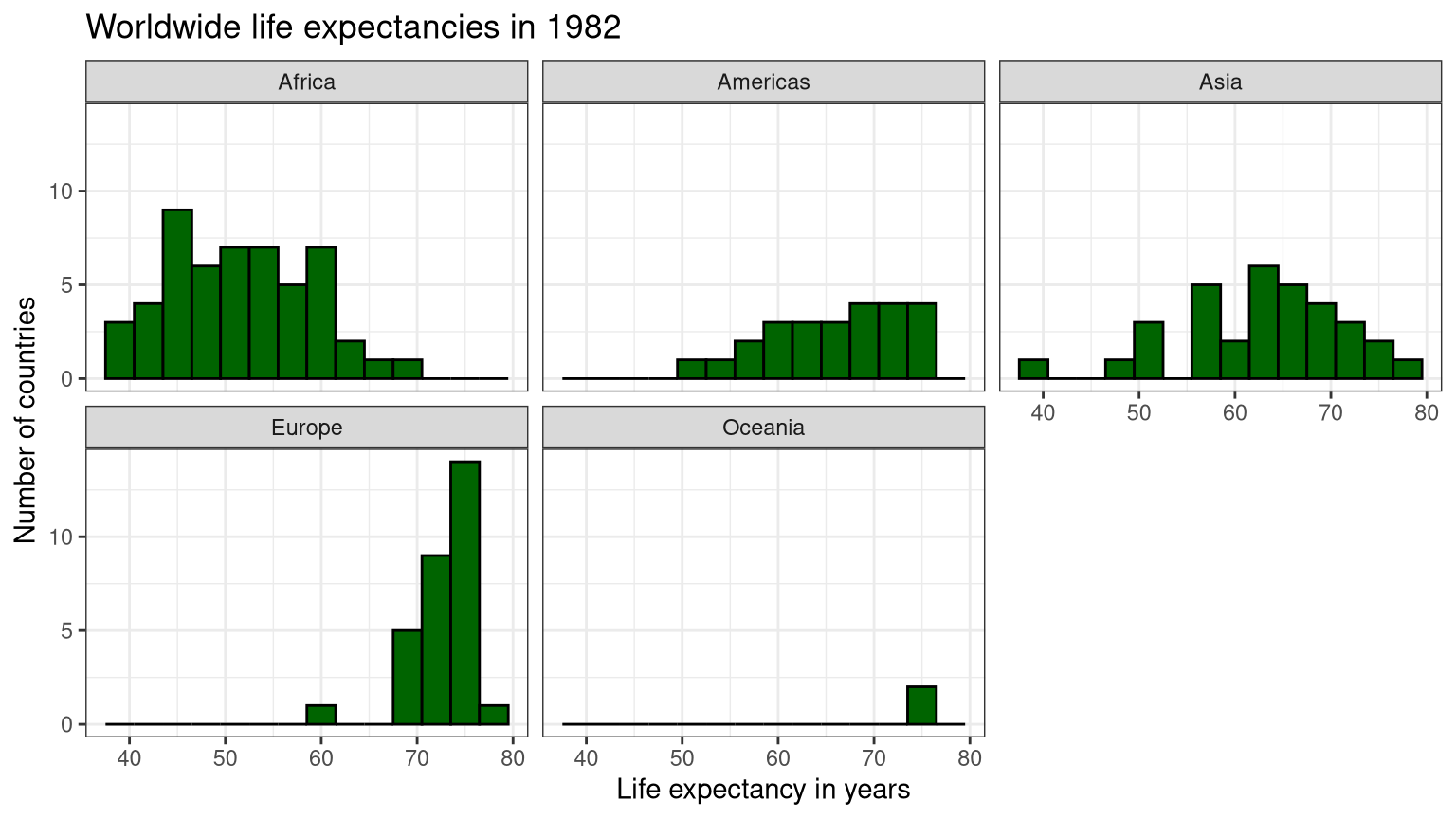
ggplot (data = gapminder1982, aes (x = lifeexp)) + geom_histogram (binwidth = 3 ,color = "black" ,fill = "darkgreen" ) + theme_bw () + labs (title = "Worldwide life expectancies in 1982" ,x = "Life expectancy in years" ,y = "Number of countries" ) + facet_wrap (vars (continent))
Code
|> group_by (continent) |> summarize (Mean = mean (lifeexp),SD = sd (lifeexp),Median = median (lifeexp),IQR = IQR (lifeexp)-> results|> :: kable ()
Table 2: Statistics by continent
Africa
51.59287
7.3759401
50.756
10.97025
Americas
66.22884
6.7208338
67.405
9.39900
Asia
62.61794
8.5352214
63.739
11.26800
Europe
72.80640
3.2182603
73.490
4.11750
Oceania
74.29000
0.6363961
74.290
0.45000
Code
<- lm (lifeexp ~ continent, data = gapminder1982)get_regression_table (lifeexp_mod) -> T2|> :: kable ()
intercept
51.593
0.955
54.051
0
49.705
53.480
continent: Americas
14.636
1.675
8.737
0
11.323
17.949
continent: Asia
11.025
1.532
7.197
0
7.996
14.054
continent: Europe
21.214
1.578
13.443
0
18.093
24.334
continent: Oceania
22.697
4.960
4.576
0
12.889
32.505
Code
term
estimate
std_error
statistic
p_value
lower_ci
upper_ci
intercept
51.593
0.955
54.051
0
49.705
53.480 continent: Americas
14.636
1.675
8.737
0
11.323
17.949 continent: Asia
11.025
1.532
7.197
0
7.996
14.054 continent: Europe
21.214
1.578
13.443
0
18.093
24.334 continent: Oceania
22.697
4.960
4.576
0
12.889
32.505
Report the average life expectancy for Africans in 1982 using lifeexp_mod.
Code
1 , "estimate" ] |> pull ()
Code
Code
The average life expectancy for Africans in 1982 was 51.593 years. Or, one could use coef(lifeexp_mod)[1] to return 51.5928654 years.
Report the average life expectancy for Europeans in 1982 using lifeexp_mod.
Code
1 ,2 ] |> pull () + T2[4 ,2 ] |> pull ()
Code
coef (lifeexp_mod)[1 ] + coef (lifeexp_mod)[4 ]
Code
round (coef (lifeexp_mod)[1 ] + coef (lifeexp_mod)[4 ],3 )
Code
predict (lifeexp_mod, newdata = data.frame (continent = "Europe" ))
The average life expectancy for Europeans in 1982 was 72.807 years.
Note: moderndive wrapper functions round answers. This is not always a good thing. It is best to leave the rounding until the very end! Consider the following inline R code.
The average life expectancy for Europeans in 1982 was 72.8064 years. Which if you want to round to three decimal places would be: 72.806 years. Another way to get the desired answer is with the predict() function. The average life expectancy for Europeans in 1982 was 72.8064 years.
Code
[1] "country" "lifeexp" "continent" "gdppercap"
Code
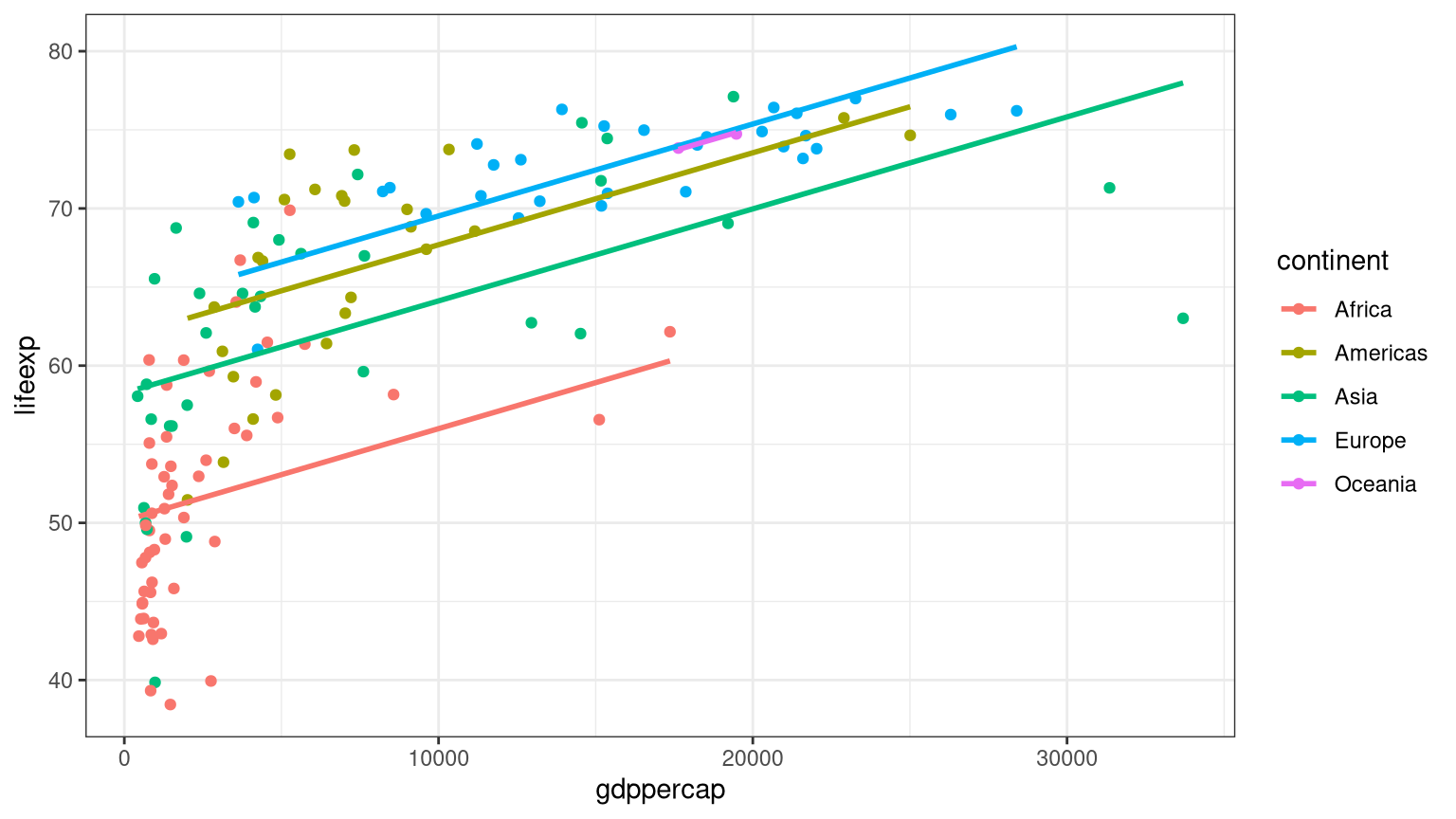
<- lm (lifeexp ~ gdppercap* continent, data = gapminder1982)<- lm (lifeexp ~ gdppercap, data = gapminder1982)anova (mod_simple, mod_full)
Analysis of Variance Table
Model 1: lifeexp ~ gdppercap
Model 2: lifeexp ~ gdppercap * continent
Res.Df RSS Df Sum of Sq F Pr(>F)
1 140 7812.3
2 132 4553.2 8 3259.1 11.81 1.358e-12 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Code
Analysis of Variance Table
Response: lifeexp
Df Sum Sq Mean Sq F value Pr(>F)
gdppercap 1 8544.6 8544.6 247.7152 < 2.2e-16 ***
continent 4 3000.6 750.1 21.7472 8.411e-14 ***
gdppercap:continent 4 258.5 64.6 1.8738 0.1187
Residuals 132 4553.2 34.5
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Code
<- lm (lifeexp ~ gdppercap + continent, data = gapminder1982)summary (modpar)
Call:
lm(formula = lifeexp ~ gdppercap + continent, data = gapminder1982)
Residuals:
Min 1Q Median 3Q Max
-18.9857 -3.0800 -0.0143 3.8538 16.6619
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 5.014e+01 8.514e-01 58.894 < 2e-16 ***
gdppercap 5.852e-04 8.495e-05 6.889 1.91e-10 ***
continentAmericas 1.170e+01 1.509e+00 7.749 1.93e-12 ***
continentAsia 8.127e+00 1.389e+00 5.851 3.48e-08 ***
continentEurope 1.353e+01 1.762e+00 7.676 2.87e-12 ***
continentOceania 1.329e+01 4.498e+00 2.955 0.00369 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.948 on 136 degrees of freedom
Multiple R-squared: 0.7058, Adjusted R-squared: 0.695
F-statistic: 65.26 on 5 and 136 DF, p-value: < 2.2e-16
Code
library (moderndive)ggplot (data = gapminder1982, aes (x = gdppercap, y = lifeexp, color = continent)) + geom_point () + geom_parallel_slopes (se = FALSE ) + theme_bw ()